Casimir Wierzynski, a senior director in Intel's AI products group, talked to me about his work in connectomics with "salami slicers" for the brain that are making it possible to map the neural networks of our minds.
CASIMIR WIERZYNSKI:
I studied electrical engineering in college and ended up on Wall Street for a while as a derivatives trader. That is, in a way, an act of AI in itself because trading is all about decision making under uncertainty and you can think of that as a nice working definition of AI, getting machines to do that instead of people. After a few years of doing that, I kind of craved going back into AI and studying it for real.
I went back to grad school to get a PhD. A really promising model for AI was the human brain itself, so I ended up, slightly to my surprise, studying neuroscience. If you want to figure out how the brain is really doing things, you have to put on a pair of gloves and start disassembling animals and poke brain cells and see what's going on in there. There are a lot of theories. I kind of describe it as theories in search of data. So to get the data I recorded from rat's brains as I taught them tasks. In the day they would learn and then at night they would sleep. I was trying to understand this connection between learning and sleep and what's the computational function of sleep. It turns out to be quite a deep topic.
After that, I got pulled into this really interesting project around neuromorphic engineering. This is the idea that you can build computer chips that actually mimic the brain. You have parts on the chip that spike and send action potentials to each other just as neurons do. And that led me back into engineering and back into how you build artificial systems that are intelligent.
The prevailing hypothesis in neuroscience is that the way the brain computes is really very much a function of its connectivity. So the connectivity form defines function. The way things are connected tells you how things work. But then that kind of begs the next question. How specifically are things connected in the brain? So that field is called connectomics. Cajal himself in the late 1800s was doing studies under a microscope and staining specific cells and following where they go. There's clearly been an interest in tracking connectivity. But from the point of view of an electrical engineer or a computer architect who actually wants to get a real detailed wiring diagram, like what specifically is connected to what, those data have been really hard to get and certainly hard to do in a systematic way.
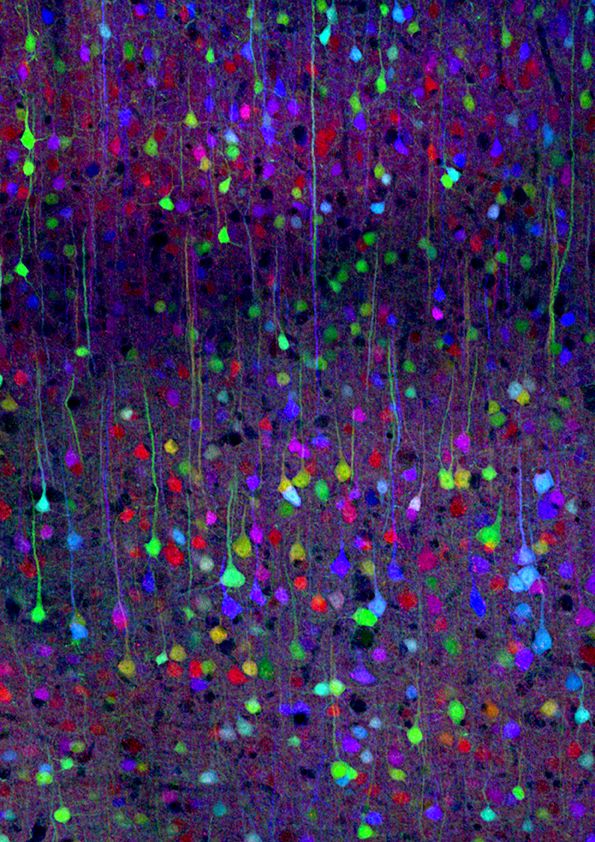
So, fast forward to around 2007, and people like Jeff Lichtman at Harvard developed these techniques, what's like an automated salami slicer for brain tissue, except these slices are 20 nanometers thick, so they wouldn’t make a great sandwich. But, this is the level of resolution that you want to resolve connections in the brain. Jeff has this amazing technology for doing the automated slicing. These little slices end up going on a plastic tape, almost like reel to reel tape from a recording studio. And then he passes this tape through an automated electron microscope machine and you end up with these super high-resolution images of successive slices of brain. That's step one. Already, that's like a heroic undertaking. But now you're left with this really interesting computational problem.
In principle, all the information about connectivity is in these slices because you can see the outlines of brain cells, the cell membranes, and you have enough resolution to see where two membranes come together, a synaptic button they call it. You even have enough resolution to see inside the synapse, the little vesicles that contain the neurotransmitter that pass chemicals from one brain cell to the other. So it's amazing resolution. If you can imagine tracing the outlines of all of these things on one slice. And then on the next slice, again, you trace all the outlines of what you saw. You could connect all of those outlines together and it would start to look like a 3-D skeleton of the neurons and the glia and all the parts of the brain. I've had the privilege, the real fun as a neuroscientist, to help on the computational side of things collaborating with Professor Nir Shavit at MIT.
How do you speed this up? Because, if it takes the age of the universe to do it, then it's not practical. So we set ourselves the goal of having the computer keep up with the microscope. That would be a great threshold because then your data don't get backlogged. I think we may be getting close to that threshold by doing clever things around systems engineering and making full use of all of the goodness in the Intel processors and so on.
So a neuron's soma, the cell body of a neuron, is typically around 20 microns. So that would be like 20,000 nanometers compared to 20 nanometers. So you build up a thousand layers to see the 3-D image of one neuron. You can create these beautiful visualizations.
One thing I think that's kind of underappreciated about the brain is just how tightly packed all of this wiring and brain cells are. What led Cajal to be able to do so much tracing of neuroanatomy just using staining and a microscope, was this technique he and Golgi figured out where only a few neurons would get stained so you could see the empty space between them. You could actually visualize what neurons look like.
So the cool thing we can do with connectomic visualizations is turn on half the brain cells and see an overall gestalt of how they're connected. But you also have the information in digital form, so you can do all the kind of cool statistics, like between this brain area and this brain area, how many brain cells on average does one cell contact. You can build up statistics like that, which you need to do science.
So, the most recent project that I saw was to slice a cubic millimeter of brain, and that took about a month. Now that's on a single slicing machine, but it did run more or less 24/7. With the human genome project, there was this initial kind of protocol for doing sequencing called Sanger sequencing. And there was some effort to get that to work on a single machine reliably. And then once they got it on the single machine, they then filled gymnasiums full of Sanger sequencing machines. So you could imagine one month for a single cubic millimeter and it kind of goes linearly from that. One day it would be 30 machines and so on.
I think mouse brain is definitely in sight within the next decade. Drosophila melanogaster, our friend the fruit fly, has basically been done at this point, at the connectomics level. So we're kind of moving our way up the evolutionary ladder.
What's interesting is that the bottleneck now is still actually the computational step, not the slicing step.
This post is adapted from the first half of Episode 32 of the podcast Eye on AI. Find the full recorded episode here, or listen to it below.